"Silent mutations" impacting cancer cells
Direktzugriff
Artikelaktionen
Gene modifications in the protein blueprint have so far been considered irrelevant if the same protein building blocks are nevertheless produced. Freiburg researchers have now shown that these mutations are able to alter the protein activity in cancer cells.
Until now, it was thought that changes in the genome would have no consequences if they do not change the protein building blocks. These are referred to as "silent mutations" or synonymous mutations. Researchers in Freiburg are now showing that such changes can, however, impact the function of the cell. The collaboration work led by Prof. Dr. Sven Diederichs, who heads the Division of Cancer Research at the Department of Thoracic Surgery at the Faculty of Medicine of the University of Freiburg and the Division of RNA Biology & Cancer - German Cancer Research Center (DKFZ), investigated 88 tumor types and more than 650,000 mutations. "Using the important cancer gene KRAS as an example, we were able to show that a supposedly silent mutation reduces protein formation. Silent mutations are therefore not so silent," said Diederichs. The researchers developed a user-friendly database, SynMICdb, which lists known synonymous mutations and thus considerably simplifies evaluation for other researchers.
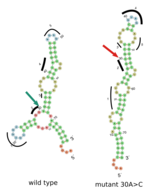
Bioinformaticians from the Department of Computer Science were able to predict misfoldings of mRNA using computational biology algorithms and big data analysis. "We were able to show that the spatial characteristics can also change considerably with silent mutations," said Prof. Dr. Rolf Backofen from the Department of Computer Science at the University of Freiburg.
The genetic code of life has redundancy. Milad Miladi, a doctoral researcher at the Department of Computer Science, explains this redundancy with the example of computer processors. "The protein-making machinery translates the 64 different RNA-triplets (biological opcodes) to only about 21 amino acids and a few production instructions. If the code of life would have been a computer machine language, then its redundancy is like having a processor architecture with multiple instruction codes for the same arithmetic operation. We have shown that even the synonymous distortions have impacts on the cells. This effect is comparable to the behavioral differences between running two logically-identical programs that have different memory fragmentation patterns and loading times."
"Such changes could play an important role in cancer therapy in the future, for example, because certain proteins are produced more or less strongly. Before this happens, however, the consequences of synonymous mutations need to be better investigated. Thanks to our online database, this is now also possible for scientists who do not have in-depth bioinformatic knowledge," said Diederichs.
Original publication:
Sharma, Y., Miladi, M., Dukare, S., Boulay, K., Caudron-Herger, M., Groß, M., Backofen, R. and Diederichs, S., 2019. A pan-cancer analysis of synonymous mutations. Nature communications, 10 no. 1 pp. 2569, 2019.
Contact:
Prof. Dr. Sven Diederichs
Division of Cancer Research
Department of Thoracic Surgery
Center for Translational Cell Research (ZTZ)
Faculty of Medicine of the University of Freiburg
Telefon: 0761 270-77571
sven.diederichs@uniklinik-freiburg.de
Prof. Dr. Rolf Backofen
Bioinformatics Group
Department of Computer Science
University of Freiburg
Telefon: 0761 203-7461
backofen@informatik.uni-freiburg.de
Fußzeile
Benutzerspezifische Werkzeuge